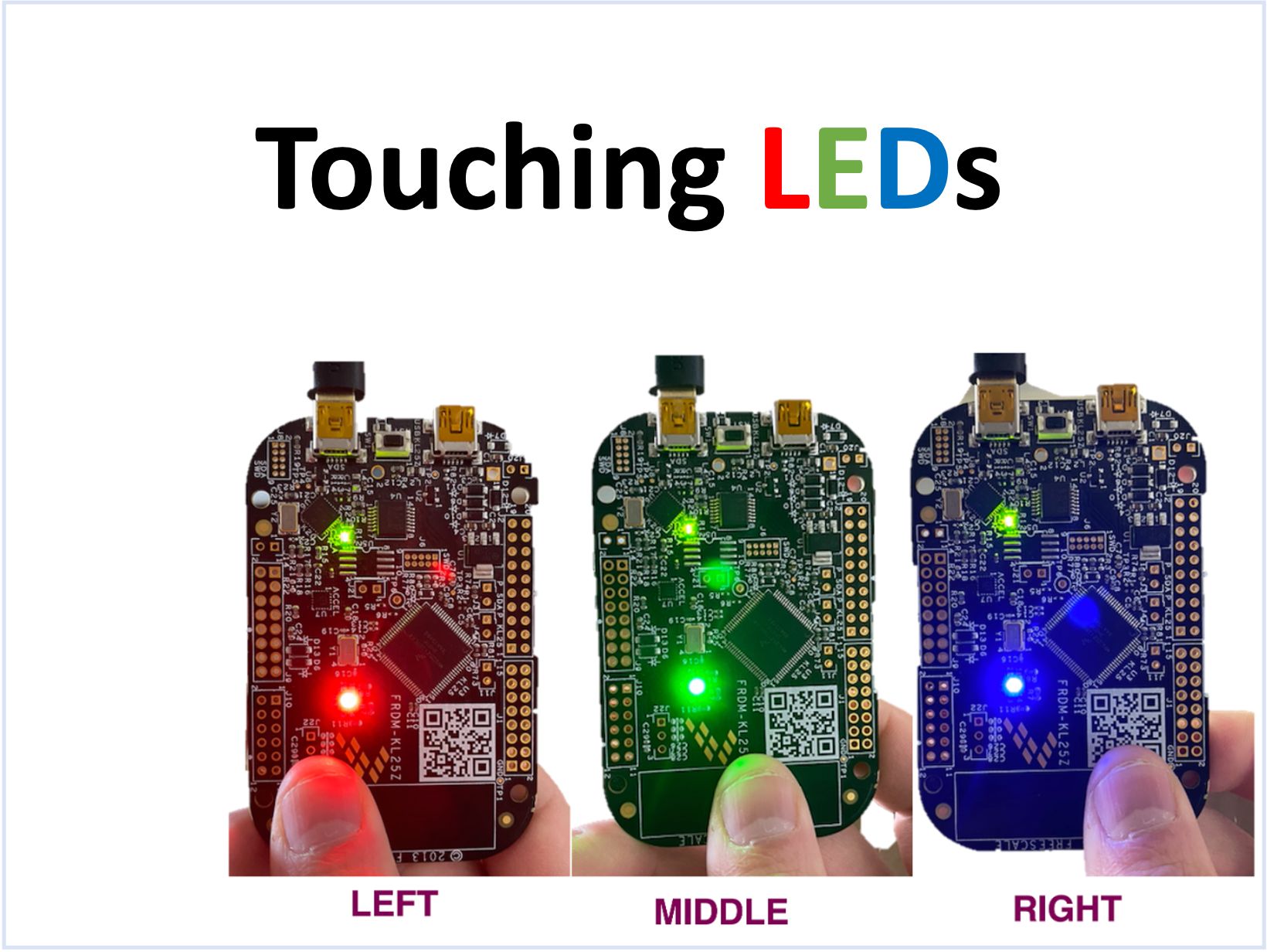
High-Level Achievements
A significant portion of my career so far has been spent developing software, ranging from bioinformatics to working with industry-level systems.
Internships/Contracts, including two interships (Microsoft and Inscripta) and one year long contract (Medtronic).
Publications primarily in the area of bioinformatics, including others that vary in subject.
Years of experience working on software tools, data analysis, and learning along the way.
Open source projects, ranging from bioinformatics to embedded systems engineering.
Primary Skillset
These areas outline where I have spent a significant portion of my time, but I am open to (and interest in) new avenues of exploration!
Clean Code (Software)
In addition to coursework, I have spent an increasing portion of time learning how to write clean code (Robert Martin ref), apply design patterns, and learn new technologies (for the latest example, Rust).
Machine/Deep Learning
Many projects I have worked on use machine learning (or AI). Likewise, I have taken courses in machine learning, data science, Robotics & AI, and deep learning.
Embedded Software
My Ph.D. focus is on embedded systems, and a large portion of my research is focused on this area.
Bioinformatics
Both my MS (Rice) and Ph.D. (CU Boulder) had projects building software with biology applications.
Skills
Coming from a background in chemistry and biology, much of the focus for the past 5 years has been on hyper-learning fundamental and advanced technologies.
Programming Languages
Backend
WebDev
Technologies
Databases
Libraries / Misc
Other
Resume
Education
MS/PhD & Computer Science (in progress)
2020 - 2023 (expected)
University of Colorado at Boulder, Boulder CO.
Thesis: “The Phage Toolbox: Automating Phage Discovery using novel software, devices, and high-throughput methodology”
Description: NSF-GRFP fellow; PhD is focused on building software and devices for automating bacteriophage research.
MS & SSPB (Bioinformatics)
2018 - 2020
Rice University, Houston TX.
Thesis: “A novel computational platform for sensitive, accurate, and efficient screening of nucleic acids”
Description: My MS at Rice was focused on both synthetic biology and computational biology.
BS & Chemistry and Biology (GPA: 3.67; 2 diplomas)
2012-2017
University of Northern Colorado, Greeley CO.
Description: Double major in chemistry and biology with graduate coursework and research internships.
Professional Experience
Microsoft: Data Scientist (PhD Intern)
Summer 2022
Seattle, WA.
-
Objective:
Investigate Evictable VMs. (Vague on Purpose) -
Key Result 1:
Contributed to the foundational codebase for the team, adding new methods for Azure-based metrics, worked with the team to create a Power BI Dashboard for collaborative insights. -
Key Result 2:
Used a multitude of ML-based approaches, gathering business-level insights into improving certain features of Spot VMs. The summer efforts resulted in an abstract submitted to Microsoft's internal MLADS conference.
Medtronic: Software/ML Engineer
Aug 2021 - May 2022
Boulder, CO.
-
Objective:
Build a device-centric machine-learning-amenable software system. -
Key Result 1:
Designed the host software system, which was able to communicate with a prototyped surgical device. (model-view-controller) -
Key Result 2:
Worked with the team to ensure code quality and maintenance, and also lead releasing the software versions. Ensured the minimal viable product contained deep learning pose prediction to assist with the device being built.
Inscripta: Computational Biologist (PhD Intern)
Summer 2021
Boulder, CO.
-
Objective:
Apply graph-genome methods towards genetic engineering. -
Key Result 1:
I used graph-genome-based algorithms, wrote objected-oriented software, and communicated findings with the scientists at Inscripta. -
Key Result 2:
Created a pipeline utilizing graph-genome approached to identify engineered genomes.
Portfolio
Here are a few of the projects I have had the oppurtunity to develop with others!
- All
- Bioinformatics
- Embedded
- Misc
Contact
Please fill free to contact me for more information. I love meeting new people, and I'm open to oppurtunities!
Dreycey Albin
Let's connect! Here are some other quick links:
Boulder, CO
albindreycey@gmail.com
+1 720 448 9778